他身在高楼广厦之中,却有山泽鱼鸟之思
532 字
3 分钟
天然氨基酸溯源
前言
TIP准备工作 分子
smiles\
:::
from rdkit import Chem
from rdkit.Chem import AllChem, DataStructs
from rdkit.Chem import Draw
from IPython.display import display
# 定义20种天然氨基酸信息
# 数据格式:单字母简写 : (英文全名, 三字母简写, 中文名称, SMILES)
amino_acids_info = {
'A': ("Alanine", "Ala", "丙氨酸", "N[C@@H](C)C(=O)O"),
'R': ("Arginine", "Arg", "精氨酸", "N[C@@H](CCCNC(N)=N)C(=O)O"),
'N': ("Asparagine", "Asn", "天冬酰胺", "N[C@@H](CC(=O)N)C(=O)O"),
'D': ("Aspartic Acid", "Asp", "天冬氨酸", "N[C@@H](CC(=O)O)C(=O)O"),
'C': ("Cysteine", "Cys", "半胱氨酸", "N[C@H](CS)C(=O)O"),
'Q': ("Glutamine", "Gln", "谷氨酰胺", "N[C@@H](CCC(=O)N)C(=O)O"),
'E': ("Glutamic Acid", "Glu", "谷氨酸", "N[C@@H](CCC(=O)O)C(=O)O"),
'G': ("Glycine", "Gly", "甘氨酸", "NCC(=O)O"),
'H': ("Histidine", "His", "组氨酸", "N[C@@H](CC1=CNC=N1)C(=O)O"),
'I': ("Isoleucine", "Ile", "异亮氨酸", "CC[C@H](C)[C@@H](N)C(=O)O"),
'L': ("Leucine", "Leu", "亮氨酸", "N[C@@H](CC(C)C)C(=O)O"),
'K': ("Lysine", "Lys", "赖氨酸", "N[C@@H](CCCCN)C(=O)O"),
'M': ("Methionine", "Met", "甲硫氨酸", "N[C@@H](CCCSC)C(=O)O"),
'F': ("Phenylalanine", "Phe", "苯丙氨酸", "N[C@@H](CC1=CC=CC=C1)C(=O)O"),
'P': ("Proline", "Pro", "脯氨酸", "N1[C@@H](CCC1)C(=O)O"),
'S': ("Serine", "Ser", "丝氨酸", "N[C@@H](CO)C(=O)O"),
'T': ("Threonine", "Thr", "苏氨酸", "N[C@@H](C(C)O)C(=O)O"),
'W': ("Tryptophan", "Trp", "色氨酸", "N[C@@H](CC1=CNC2=CC=CC=C12)C(=O)O"),
'Y': ("Tyrosine", "Tyr", "酪氨酸", "N[C@@H](CC1=CC=C(O)C=C1)C(=O)O"),
'V': ("Valine", "Val", "缬氨酸", "N[C@@H](C(C)C)C(=O)O")
}
def print_compare_smiles(smiles_list, legends=None):
"""
通用显示函数,可以接受一个包含4个SMILES的列表进行绘图比较。
"""
mols = [Chem.MolFromSmiles(s) for s in smiles_list]
if any(m is None for m in mols):
print("无效的SMILES字符串存在")
return
for m in mols:
AllChem.Compute2DCoords(m)
if legends is None:
legends = ['Mol1', 'Mol2', 'Mol3', 'Mol4']
# 显示4个分子,2x2网格
img = Draw.MolsToGridImage(
mols,
legends=legends,
subImgSize=(300,300),
molsPerRow=2,
returnPNG=False
)
display(img)
def get_top3_similar_amino_acids(query_smiles):
query_mol = Chem.MolFromSmiles(query_smiles)
if query_mol is None:
print("查询SMILES无效。")
return None
query_fp = AllChem.GetMorganFingerprintAsBitVect(query_mol, 2, nBits=1024)
similarities = []
for one_letter, info in amino_acids_info.items():
name_en, name_3, name_cn, smiles = info
aa_mol = Chem.MolFromSmiles(smiles)
if aa_mol:
aa_fp = AllChem.GetMorganFingerprintAsBitVect(aa_mol, 2, nBits=1024)
sim = DataStructs.TanimotoSimilarity(query_fp, aa_fp)
similarities.append((one_letter, name_en, name_3, name_cn, smiles, sim))
# 根据相似度降序排序
similarities.sort(key=lambda x: x[5], reverse=True)
return similarities[:3]
# 使用示例
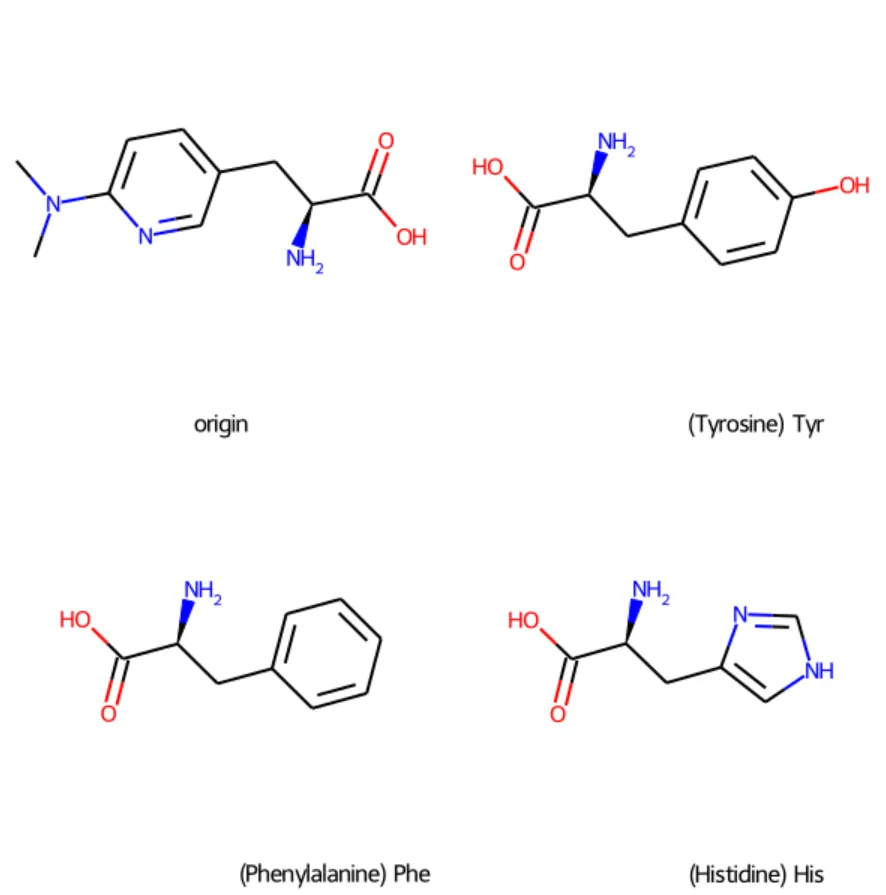
query_smiles = "CN(C)c1ccc(C[C@H](N)C(=O)O)cn1"
#query_smiles = "CC[C@@H](N[C@H](c1nc(C(=O)O)cs1)C(C)C)c1nnc([C@@H](N)Cc2ccccc2)o1"
#CC[C@@H](N[C@H](c1nc(C(=O)O)cs1)C(C)C)c1nnc([C@@H](N)Cc2ccccc2)o1
top3 = get_top3_similar_amino_acids(query_smiles)
if top3 and len(top3) == 3:
print("与查询分子最相似的三个氨基酸及其相似度:")
for i, t in enumerate(top3, start=1):
one_letter, name_en, name_3, name_cn, smiles, sim = t
print(f"排名{i}: {name_cn}({name_en}), 三字母:{name_3}, 单字母:{one_letter}, SMILES:{smiles}, 相似度={sim:.3f}")
# legends中包含原始分子、以及top3氨基酸的标注,标注形式:中文名(英文名)三字母简写
legends = ["origin"] + [f"{x[3]}({x[1]}) {x[2]}" for x in top3]
smiles_to_show = [query_smiles] + [x[4] for x in top3]
print_compare_smiles(smiles_to_show, legends=legends)
图片参考 smiles